Spatial Transcriptomics measures the total mRNA of a complete tissue section, combines the spatial information of the total mRNA with its morphological content, and maps the locations of all gene expression to obtain a complex and complete gene expression map of biological processes. As this technology determines different cell populations while preserving their spatial location, it provides important information for the relationship between cell function, phenotype, and location in the tissue microenvironment. It can reveal signal pathways activated in fine physiological regions, and complete the analysis of the mechanism of molecular characteristics driving biological characteristics. Grandomics provides spatial transcriptome analysis services based on the NanoString platform with extremely high sensitivity, accuracy and repeatability. This technology does not require enzymatic reaction, reverse transcription, and PCR amplification, further reducing the generation of errors. Therefore, nCounter has an unparalleled advantage in the field of expression profile quantitative analysis.
Product Advantages
As a new generation of molecular labeling technology, NanoString technology detaches itself from the principles of traditional PCR technology, expression profile chip technology and RNA-Seq technology based on second-generation sequencing. It does not require reverse transcription, amplification, and technical duplication. The target nucleic acid in the sample is subjected to quantitative analysis, with little interference from external sources, ensuring the accuracy of the results. This technology has unique advantages in the RNA quantification of paraffin sections. The specially designed 50nt gene-specific hybridization sequence greatly reduces the interference caused by the RNA degradation of clinical samples. It can also achieve accurate quantification in RNA degradation samples (low RIN value), and has better repeatability than PCR in clinical sample analysis, which maximizes the effective information of clinical samples.
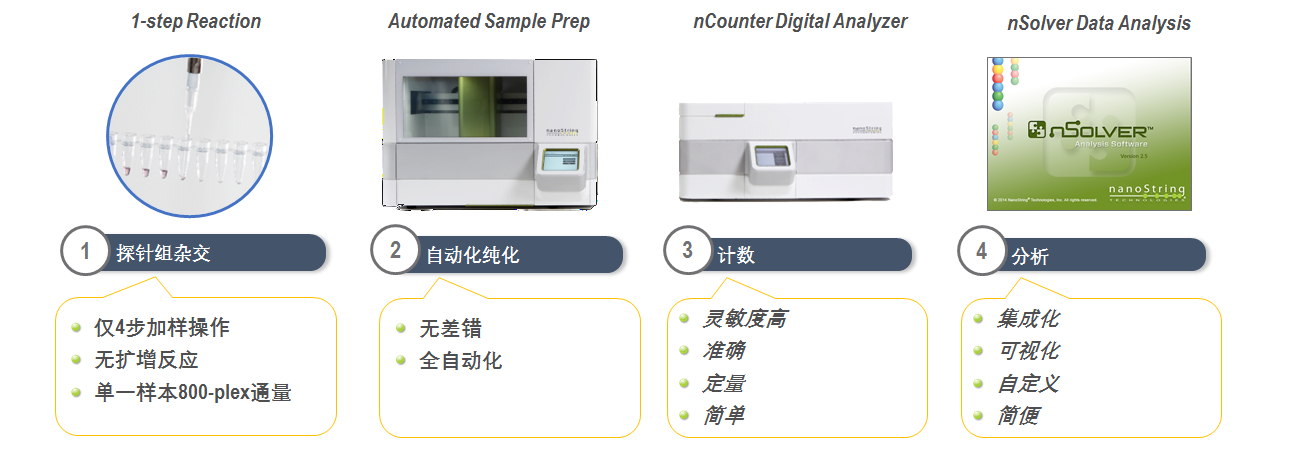
Technical workflow
Experiment Content
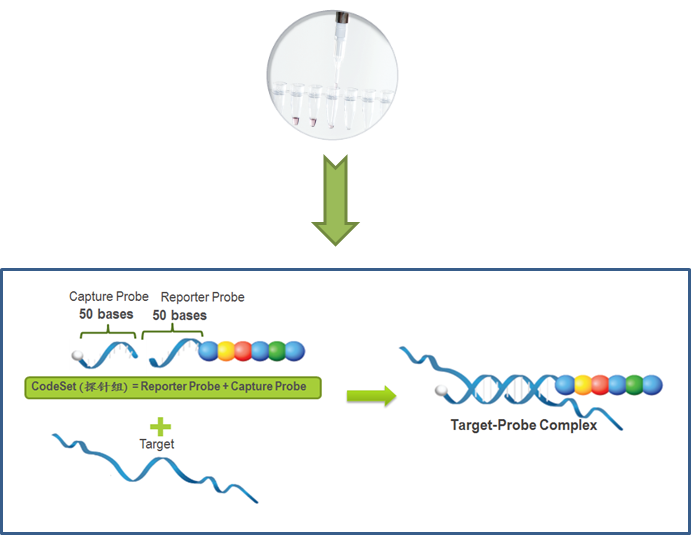
1.Hybridization of fluorescent coding probe with target molecule
The fluorescent coding probe has two parts
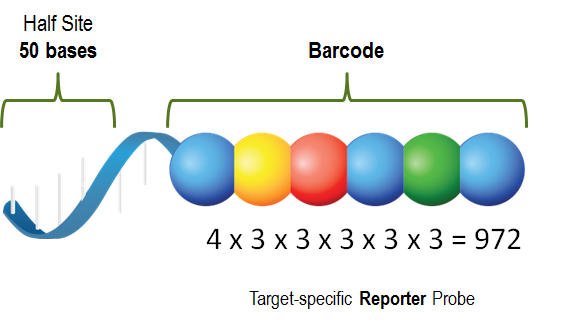
- Reporter Probe is a 50-mer nucleic acid sequence specific to the target molecule, coupled with different fluorescent color combinations (CodeSet). It is the main source of the detection signal. Fluorescent probes can achieve 800 different permutations and combinations to achieve the effect of distinguishing different target molecules.
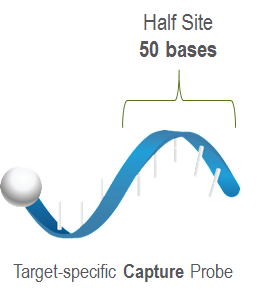
- Capture Probe: It is a probe with a 50 mer nucleic acid sequence specific to the target molecule, coupled with biotin. Its purpose is to make the probe capture the target molecule and fix it on the avidin-coated cartridge
- Molecular hybridization: This process is the process of hybridizing the reporter probe, the capture probe and the target molecule mixture (sample) in the EP tube. After hybridization, various target molecule-specific complexes (Target-Probe Complex) are formed in the liquid phase. ). On Cartridge coated with avidin.
2.Purification and immobilization of target specific complex
After the hybridization is completed, the sample is sent to the fully automated Prep Station to remove the remaining hybridized probes, and the purified target molecule complex is joined to the cartridge coated with avidin (by a biotin-avidin binding reaction ) and electrified to align them in the same direction. 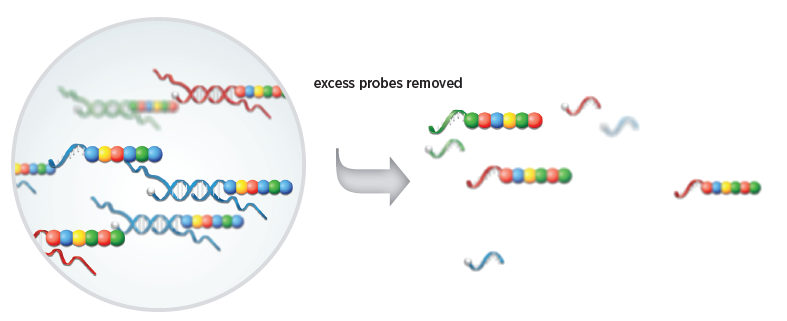
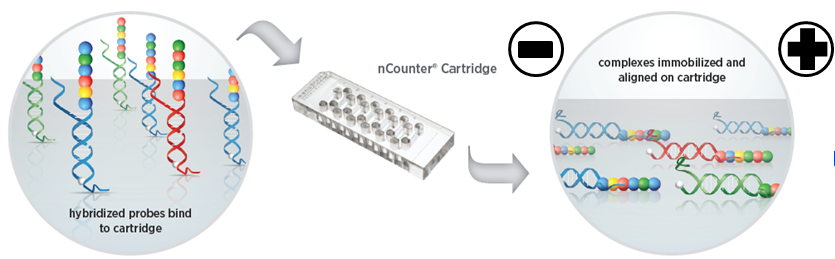
3.Signal acquisition and quantification
Move the cartridges to the nCounter Digital Analyzer for signal data collection and count. Each count corresponds to the number of specific fluorescent coding probes (one molecule = one count) on the cartridge surface. These are marked in a list, and the final quantitative statistics is performed to determine the number of all target molecules in the reaction system. 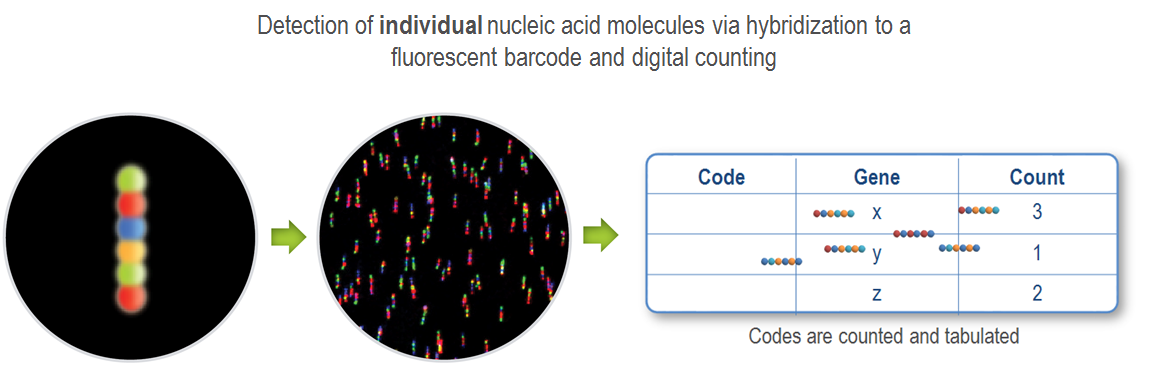
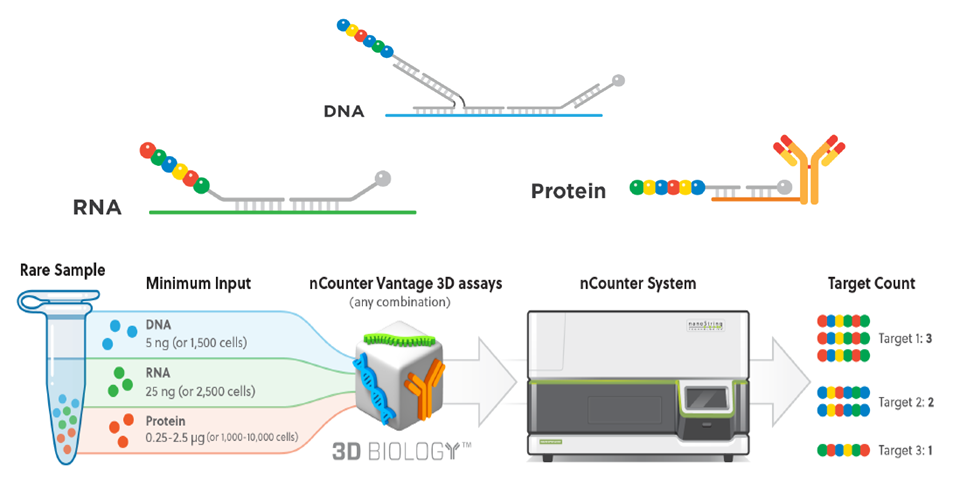
The nanoString nCounter technology can realize multi-omics co-detection of DNA, RNA and protein on the same sample. A total of 30 protein and 770 RNA expression profiles can be analyzed. This technology eliminates the differences between different analyses, and realizes 3D omics analysis in a true sense. The company’s chief scientist, Dr. Gordon Mills, as one of the co-inventors of this technology, also conducted in-depth cooperation with nanoString in the early stage of technology development to jointly develop this technology in the field of tumor gene expression profiling and protein profile signaling pathways.