Product advantages
The pan-genome uses high-throughput sequencing and biological information analysis methods to sequence and analyze individuals of different subspecies that are inherently related. After separate assembly, multiple genomes are obtained, allowing the refinement of the species gene set and the compilation of individual-specific DNA sequences and functional gene information. Such information is helpful for understanding the molecular evolutionary mechanism of speciation and its relationship with natural selection.
Strategy
| Species Type | Simple Genome | Complex Genome |
| Size<2Gb Heterozygosity<0.5% Repetetive Sequence Ratio<50% | Size>2Gb Heterozygosity>0.5% Repetetive Sequence Ratio>50% | |
| Library Type | Nanopore Ligation 1D Library Nanopore Ultra-long reads Library PacBio CLR Library PacBio HiFi reads Library | |
| Sequencing Strategy | Recommended strategy for fine mapping: 100X Nanopore/PacBio CLR+50X NGS; Higher assembly index can be obtained by combining Nanopore Ultra-long reads, PacBio HiFi reads, and Bionano and HiC technologies Other draft recommendations: 50X Nanopore/PacBio CLR | Depending on specific species |
| Fine Mapping Commitment Index | Contig N50>1Mb | Depending on specific species |
Analysis
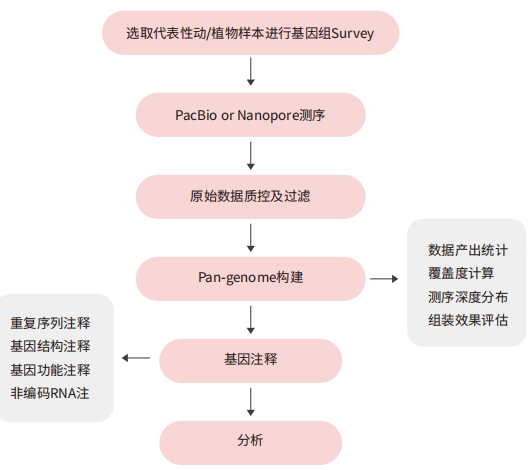
Case Analysis
Case Analysis
Case 1 Nature Genetics‖ 725 tomato pan-genomes reveal genetic changes during tomato domestication
Based on the genome sequences of 725 representative tomato germplasms, this study used a’map-to-pan’ strategy to construct a tomato pan-genome, capturing 4,873 protein-coding genes that do not exist in the reference genome, revealing differences in the presence/absence variation (PAV) of important functioning genes caused by selection. Simultaneously, researchers also identified a rare allele TomloxC caused by a promoter change, and analyzed its changes during domestication. The pan-genome increases the depth and integrity of the tomato reference genome and is of great significance for future biological discovery and breeding.
The pan-genome contains 351Mb sequences, including 372 SLL (S. lycopersicum var. lycopersicum), 267 SLC (S. lycopersicum var. cerasiforme), which are cultivated tomatoes, and 78 SP (Solanum pimpinellifolium) and 8 SCG (S. cheesmaniae and S. galapagense), which are wild tomatoes. It was observed that 25.8% of the genes showed varying degrees of PAV, indicating that at least part of the decrease in genetic diversity can be attributed to subgene loss during domestication and improvement.
The selection of promoter regions that affect downstream gene expression has also resulted in improvements in tomato domestication and genetic results. The strong negative selection of the non-reference alleles of the TomLoxC promoter during the domestication period indicates that modern tomato breeding focuses overly on traits such as yield, preservation period, and resistance to biotic and abiotic stresses, and often overlooks sensory traits and flavor quality characteristics that are hard to select, leading to the reduction of flavor-related volatiles.
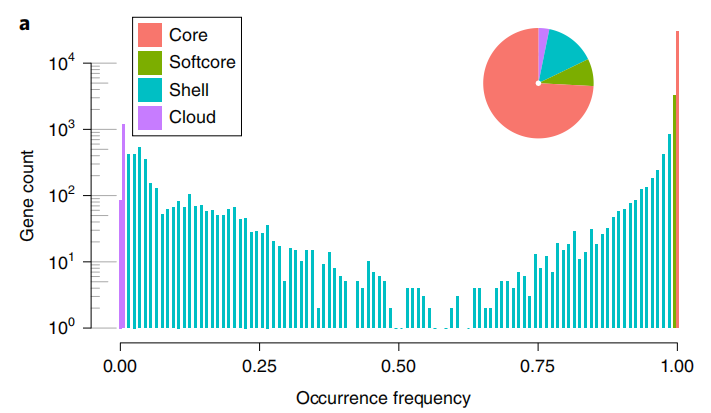
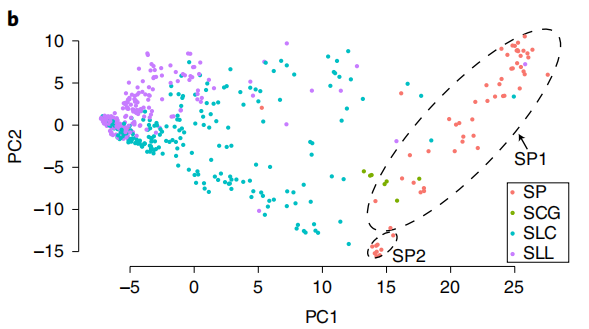
图1 番茄泛基因组组成及PAV主成分分析
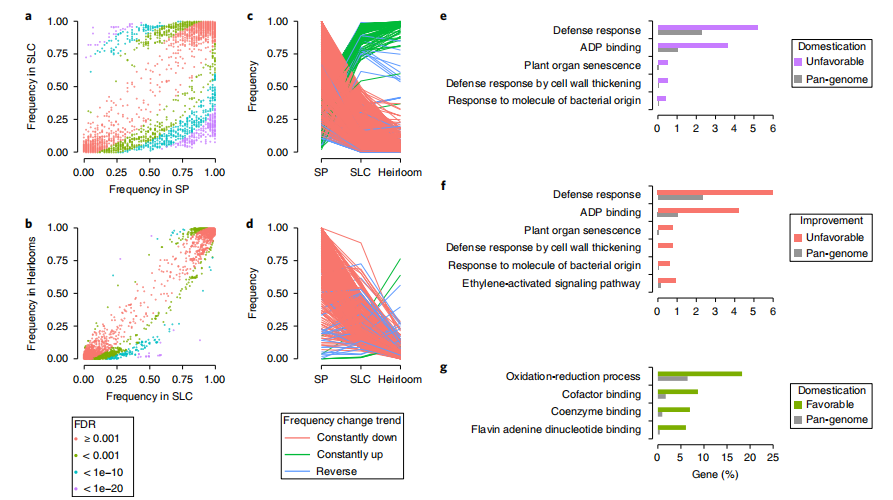
图2 番茄驯化改良过程中的基因选择偏好



