Single cell transcriptome
DNBe lab C4 single cell transcriptome sequencing
DNBe lab C4 single cell transcriptome sequencing
Product Advantages
1.Detects the expression of important single-cell genes that cannot be accurately measured with other methods due to cell heterogeneity. In samplessuch as tissue and blood, the cell composition in the sample maynot be singular, or a certain type of dominant cell population may have higher gene expression. This masks the gene expression of a small number of cell populations. Conventional RNA mixed pool sequencing (Bulk sequencing) cannot accurately determine the gene expression level of a certain type of cell population.
2.The DNBe lab C4 single cell system can label 1000-4000 cells each time. According to customer needs, multiple labeling can be performed to obtain a large number of labeled single cell populations. The beads contain more than 107 capture sequencesthat have highcapture efficiency. At the same time, due to the device’s portability, it is easily operable and there is no need for power startup and installation. This greatly reduces the cost of a single library.
Strategy
Sample requirements: single cell suspension
Target cell number: 1000-4000 cells/run, starting cell volume 1×105 cells/run
Cell diameter: 5um-25um
Sequencing data volume: 100K reads/cell
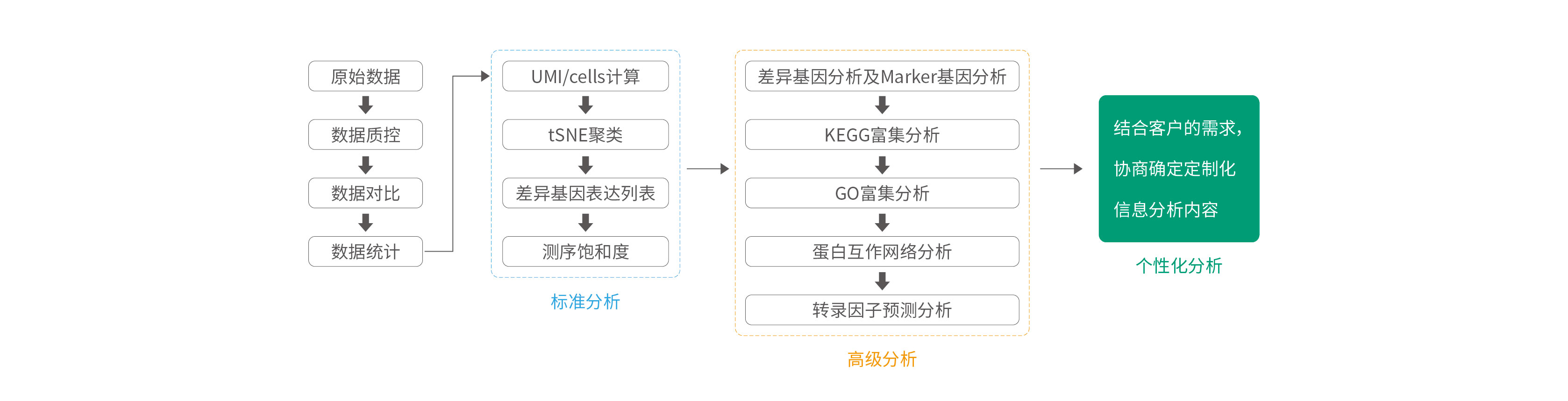
Analysis
Common Questions
1. If I only isolate 2000 cells on site with a high cell viability rate, can I directly use 2000 cells on the DNBe lab C4 platform?
Answer: Not recommended. Because the DNBe lab C4 single-cell RNA-Seq requires at least 10^5 cells each time, the cell viability is recommended to be above 80%, and the initial sample delivery volume is recommended to be at least 10^6 cells.
2.Since it is single-cell sequencing, why should the initial sample delivery volume be more than 10^6 cells?
Answer: After the sample enters the experiment center, there will be steps to remove dead cells and test cell viability. Each step will cause cell loss. Therefore, it is recommended to ensure that the sample volume is relatively large.
Case Analysis
Application of DNBelab C4 Single Cell Sequencing in Cell Atlas
The study used the DNBelab C4 portable single-cell system to perform single-cell transcriptome sequencing on the main tissues and organs of cynomolgus monkeys (lung, kidney, liver, pancreas, brain, aorta, thyroid, parotid gland and blood), and established a single-cell transcriptome for each, identifying 44 main cell types of cynomolgus monkeys. Researchers completed the construction of the first non-human primate single cell map, which has an important guiding role in the development of coronavirus drugs and vaccines.

Cynomolgus monkey tissue and organ cell map
Single-cell RNA-seq reveals 9 types of organ cell maps for cynomolgus monkeys
Through the C4 system, single cell/single cell nuclear suspension of each tissue and organ of the cynomolgus monkey was prepared into labeled single cells. Second-generation transcriptome sequencing was performed. Using the obtained single cell data of 9 tissues and organs, a total of 44 cell types were obtained through UMAP analysis.
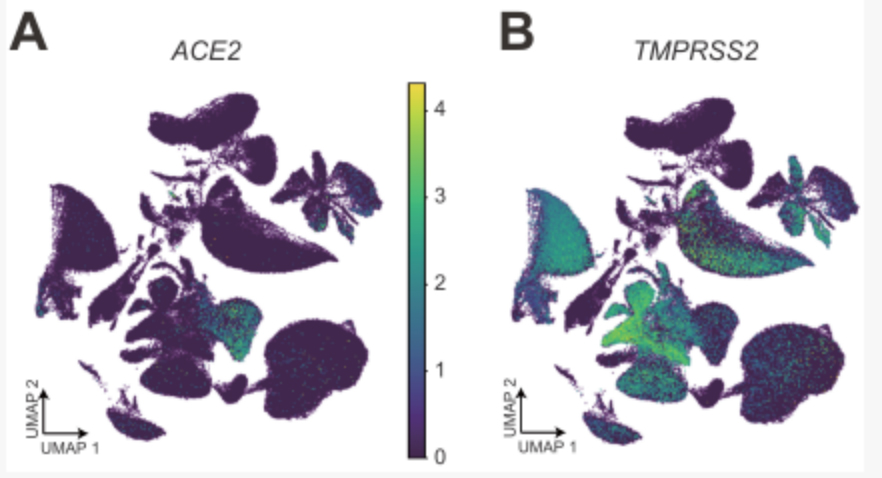
The expression of A&B ACE2 and TMPRSS2 in cynomolgus monkey tissues and organ cell groups
Further analysis of the co-expression of ACE2 and TMPRSS2 and subsequent analysis found that a kind of epithelial cell club cell in the lung is the cell type with the highest proportion of ACE2 and TMPRSS2 double-positive cells, suggesting that this type of lung epithelial cell is extremely vulnerable to coronavirus.
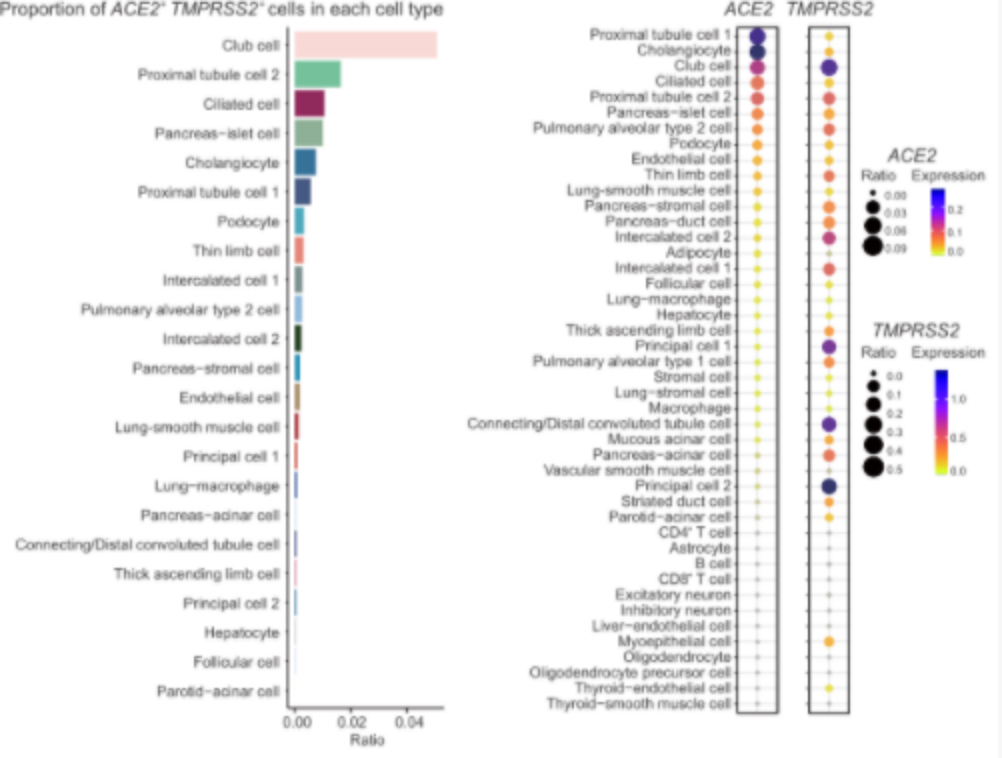
Tissue distribution of coronavirus receptor expression
Compare the expression of ACE2 and TMPRSS2 in the single-cell transcriptome data of human and cynomolgus monkey
After comparative analysis with human single-cell data, it was found that the expression of TMPRSS2 in the cynomolgus monkey kidney, lung and liver is similar to that of human corresponding organs. Alternatively, the expression of ACE2 in the cynomolgus monkey kidney, lung and liver is different from corresponding human organs. ACE2 is expressed highest in ciliated cells of cynomolgus monkey lungs, and highest in human alveolar type 2 cells. Among the proximal tubular epithelial cells of the kidney, ACE2 is expressed the highest in humans and cynomolgus monkeys. In liver tissues, the expression of ACE2 in bile duct cells is similar to that in humans and cynomolgus monkeys, while the expression level of ACE2 in human liver endothelial cells is higher than that of the same type of cells in cynomolgus monkeys. These results show the comparison of susceptibility to coronavirus between cynomolgus monkey cells and human cells, providing guidance for animal experiments and mechanism studies of new coronavirus drugs and vaccines in non-human primates.
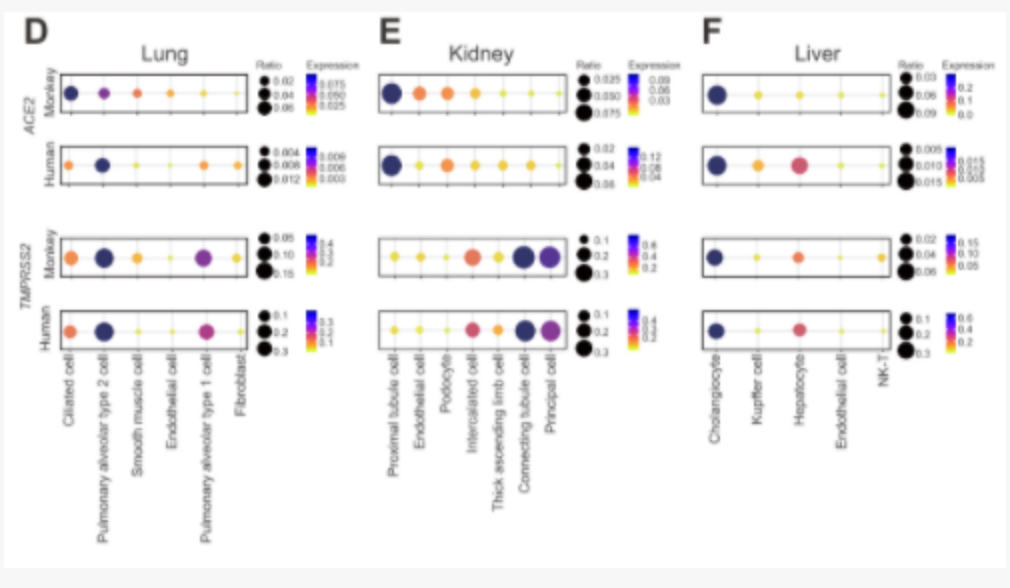
Comparison and analysis of transcription differences between human and monkey coronavirus receptor ACE2 and TMPRSS2 in lung, kidney and liver
The epigenetic regulation of ACE2 in cynomolgus monkey kidney cells
Through ACE2 expression-related gene analysis and screening, a series of highly correlated genes were discovered, including some clinically related drug targets related to immune cell failure, providing an important basis for clinical drug treatment. At the same time, the DNBe lab C4 system was used to perform chromatin accessibility sequencing (ATAC-seq) of cynomolgus monkey liver single cells. Through the integration of single-cell scATAC-seq and scRNA-seq data, it is found that the interferon-responsive cis-element (IRF1) and the binding site of the STAT transcription factor are enriched near the transcription start site of ACE2, which can connect the expression of ACE2 to the severe immune response of some patients. In particular, it was found that the core inflammatory factor IL-6 receptor, IL-6R, and ACE2 in the clinical inflammatory factor storm have co-expression patterns in tissues, suggesting that the positive feedback regulation of IL6 and ACE2 may be an important cause of the inflammatory factor storm observed clinically.
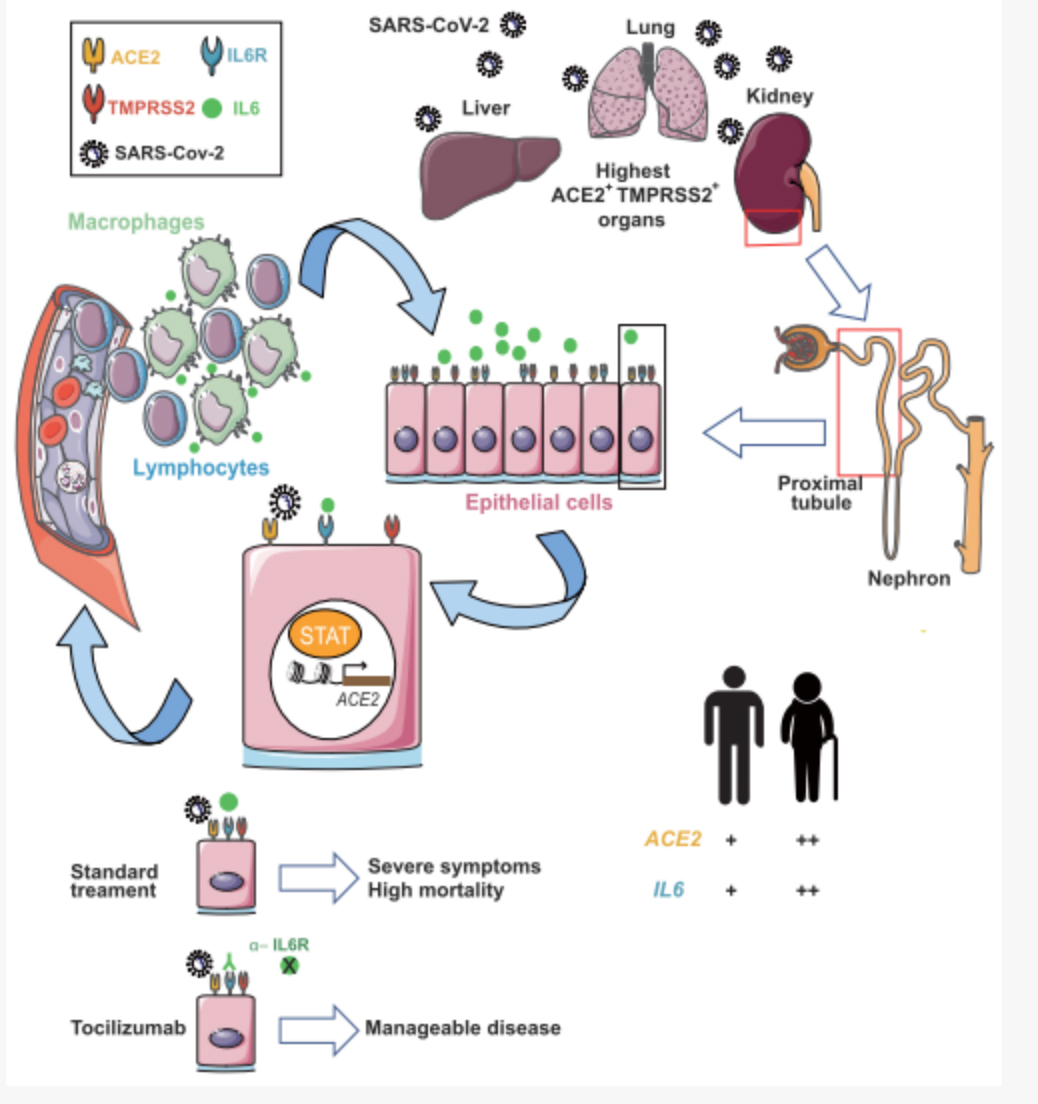
New coronavirus infection initiates IL-6 immune signaling pathway mechanism
References:
Lei Han, Xiaoyu Wei, Chuanyu Liu, et al. Single-cell atlas of a non-human primate reveals new pathogenic mechanisms of COVID-19. BioRxiv 2020.04.10.022103; doi: https://doi.org/10.1101/2020.04.10.022103.